Abstract
Introduction
Frontline treatment of patients in chronic phase of chronic myeloid leukemia(CP-CML) is classically based on tyrosine kinase inhibitors (TKI). Imatinib was introduced in 1998, then second- and third-generation TKIs have been developed. This therapeutic arsenal suggest a possible personalized treatment. The choice of TKI could be guided on the one hand by the potential adverse effects depending on the co-morbidities and on the other hand, the efficiency of treatment since an optimal response during the first year is a therapeutic goal (Baccarani M et al. A review of the European LeukemiaNet recommendations for the management of CML. Ann Hematol 2015, 94 Suppl 2:S141-7). Treatment inefficacy may be related to the persistence of quiescent leukemic cells, characterized by a decreased metabolic activity and redox metabolism inducing a low proliferation rate athough the BCR-ABL mutation is present. The level of reactive oxygen species (ROS) being regulated by antioxidant enzymes, we hypothesized that scoring gene expression levels of major antioxidant enzymes could be of clinical interest when considering the response to imatinib. By combining the gene expression profiles of CP-CML patients at the time of diagnosis and their molecular response (BCR-ABL1/ABL1 ratio) at one-year, we determined a theranostic Imatinib-Response Score (IRS) potentially useful to optimize therapeutic decisions (patent # WO2016083742).
Methods
The expression of antioxidant genes was quantified from blood samples collected from 122 CP-CML patients at diagnosis and 102 healthy volunteers (HealthOx protocol, ClinicalTrials.gov # NCT02789839). Sokal-scores were calculated and all patients were treated with imatinib for at least one year. Quantification of the BCR-ABL1/ABL1 ratio allowed to determine of the one-year response according to the ELN definition (optimal: BCR-ABL1/ABL1 ≤ 0.1%, warning: BCR-ABL1/ABL1 0.1-1%, failure: BCR-ABL1/ABL1 > 1%). RNA extraction was performed after red blood cell lysis and RNA quality was checked with a 2100 Bioanalyzer (Agilent). The expression of antioxidant genes (SOD1, SOD2, SOD3, CAT, TXN, TXN2, GLRX, GLRX2, GLRX3, GLRX5, GPX1, GPX2, GPX3, GPX4, GPX7, GSR, PRDX1, PRDX2, PRDX3, PRDX4, PRDX5, PRDX6) and 3 housekeeping genes (ACTB, B2M, RPL13A) was quantified by RT-qPCR (LightCycler® 480 and UPL technology, Roche). All targets were concomitantly analyzed in triplicates and average values from patients and aged-matched healthy controls were used to determine Relative Quantification (RQ) values by the 2-ΔΔCt method. Mutations in the ABL kinase domain were studied by direct sequencing. IRS were determined by logarithmic logistic regression performed thanks to the glm() function of the stats package (R v3.2.2 software). The generalized linear model was obtained by logistic regression with weighted RQ and was validated by split-sample strategy (10,000 repeats). Samples were divided into two groups ("learning" and "test" groups) randomly mixing optimal responses and failures.
Results and conclusion
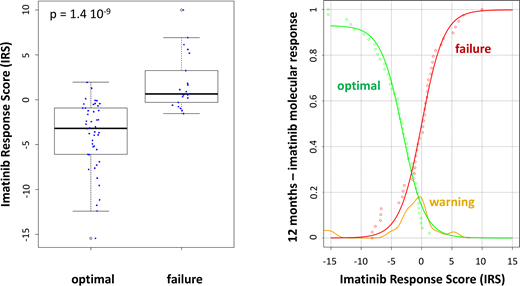
None of the patients had a mutation in the BCR-ABL kinase domain. The expression levels of numerous antioxidant genes were different when considering optimal response and failure to imatinib treatment. A multifactorial strategy by linear combination of normalized RQ values was used to calculate IRS, which allowed for an efficient discrimination between optimal response(IRS = -3.42±1.44) and failure (IRS = 1.76±0.64) (p-value = 1.4 10-9). The probability of one-year response to imatinib was assessed by empirical cumulative distribution function. This function allowed the prediction of optimal response and failure. As expected, the IRS values of patients with a warning response, not used to build the mathematical model (external validation), were located between those of optimal response and failure. Interestingly, the IRS was not correlated with Sokal-score of CP-CML patients (R2= 0.0333), reinforcing its potential usefulness for clinical management.
Altogether, this retrospective multicentric study allowed the determination of a molecular score to predict imatinib response. This simple biological strategy using diagnosis blood sample of CP-CML patients will benefit from prospective studies to adapt therapy.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal